Spatial Operations
This notebook shows how to do some spatial operations. We focus here on averaging, since the sums can easily be infered.
[1]:
import xarray as xr # requires >= 0.15.1
import numpy as np
[2]:
import warnings
warnings.filterwarnings("ignore")
[3]:
%matplotlib inline
This is where you can find the dataset on the opendap (unfortunately parallel computing with dask won’t work)
dataurl = 'http://35.188.34.63:8080/thredds/dodsC/OM4p5/'
ds = xr.open_dataset(f'{dataurl}/ocean_monthly_z.200301-200712.nc4',
chunks={'time':1, 'z_l': 1}, engine='pydap')
I am using my local copy:
[4]:
dataurl = '/work/Raphael.Dussin/runs/OM4p5_sample/'
ds = xr.open_dataset(f'{dataurl}/ocean_monthly_z.200301-200712.nc4',
chunks={'time':1, 'z_l': 1}, engine='netcdf4')
[5]:
ds
[5]:
- nv: 2
- time: 60
- xh: 720
- xq: 720
- yh: 576
- yq: 576
- z_i: 36
- z_l: 35
- nv(nv)float641.0 2.0
- long_name :
- vertex number
- units :
- none
- cartesian_axis :
- N
array([1., 2.])
- xh(xh)float64-299.8 -299.2 ... 59.25 59.75
- long_name :
- h point nominal longitude
- units :
- degrees_east
- cartesian_axis :
- X
array([-299.75, -299.25, -298.75, ..., 58.75, 59.25, 59.75])
- xq(xq)float64-299.5 -299.0 -298.5 ... 59.5 60.0
- long_name :
- q point nominal longitude
- units :
- degrees_east
- cartesian_axis :
- X
array([-299.5, -299. , -298.5, ..., 59. , 59.5, 60. ])
- yh(yh)float64-77.91 -77.72 ... 89.68 89.89
- long_name :
- h point nominal latitude
- units :
- degrees_north
- cartesian_axis :
- Y
array([-77.907938, -77.723813, -77.539688, ..., 89.472 , 89.6832 , 89.8944 ]) - yq(yq)float64-77.82 -77.63 -77.45 ... 89.79 90.0
- long_name :
- q point nominal latitude
- units :
- degrees_north
- cartesian_axis :
- Y
array([-77.815875, -77.63175 , -77.447625, ..., 89.5776 , 89.7888 , 90. ]) - z_i(z_i)float640.0 5.0 15.0 ... 6.25e+03 6.75e+03
- long_name :
- Depth at interface
- units :
- meters
- cartesian_axis :
- Z
- positive :
- down
array([0.000e+00, 5.000e+00, 1.500e+01, 2.500e+01, 4.000e+01, 6.250e+01, 8.750e+01, 1.125e+02, 1.375e+02, 1.750e+02, 2.250e+02, 2.750e+02, 3.500e+02, 4.500e+02, 5.500e+02, 6.500e+02, 7.500e+02, 8.500e+02, 9.500e+02, 1.050e+03, 1.150e+03, 1.250e+03, 1.350e+03, 1.450e+03, 1.625e+03, 1.875e+03, 2.250e+03, 2.750e+03, 3.250e+03, 3.750e+03, 4.250e+03, 4.750e+03, 5.250e+03, 5.750e+03, 6.250e+03, 6.750e+03]) - z_l(z_l)float642.5 10.0 20.0 ... 6e+03 6.5e+03
- long_name :
- Depth at cell center
- units :
- meters
- cartesian_axis :
- Z
- positive :
- down
- edges :
- z_i
array([2.5000e+00, 1.0000e+01, 2.0000e+01, 3.2500e+01, 5.1250e+01, 7.5000e+01, 1.0000e+02, 1.2500e+02, 1.5625e+02, 2.0000e+02, 2.5000e+02, 3.1250e+02, 4.0000e+02, 5.0000e+02, 6.0000e+02, 7.0000e+02, 8.0000e+02, 9.0000e+02, 1.0000e+03, 1.1000e+03, 1.2000e+03, 1.3000e+03, 1.4000e+03, 1.5375e+03, 1.7500e+03, 2.0625e+03, 2.5000e+03, 3.0000e+03, 3.5000e+03, 4.0000e+03, 4.5000e+03, 5.0000e+03, 5.5000e+03, 6.0000e+03, 6.5000e+03]) - time(time)object2003-01-16 12:00:00 ... 2007-12-16 12:00:00
- long_name :
- time
- cartesian_axis :
- T
- calendar_type :
- NOLEAP
- bounds :
- time_bnds
array([cftime.DatetimeNoLeap(2003, 1, 16, 12, 0, 0, 0, 2, 16), cftime.DatetimeNoLeap(2003, 2, 15, 0, 0, 0, 0, 4, 46), cftime.DatetimeNoLeap(2003, 3, 16, 12, 0, 0, 0, 5, 75), cftime.DatetimeNoLeap(2003, 4, 16, 0, 0, 0, 0, 1, 106), cftime.DatetimeNoLeap(2003, 5, 16, 12, 0, 0, 0, 3, 136), cftime.DatetimeNoLeap(2003, 6, 16, 0, 0, 0, 0, 6, 167), cftime.DatetimeNoLeap(2003, 7, 16, 12, 0, 0, 0, 1, 197), cftime.DatetimeNoLeap(2003, 8, 16, 12, 0, 0, 0, 4, 228), cftime.DatetimeNoLeap(2003, 9, 16, 0, 0, 0, 0, 0, 259), cftime.DatetimeNoLeap(2003, 10, 16, 12, 0, 0, 0, 2, 289), cftime.DatetimeNoLeap(2003, 11, 16, 0, 0, 0, 0, 5, 320), cftime.DatetimeNoLeap(2003, 12, 16, 12, 0, 0, 0, 0, 350), cftime.DatetimeNoLeap(2004, 1, 16, 12, 0, 0, 0, 3, 16), cftime.DatetimeNoLeap(2004, 2, 15, 0, 0, 0, 0, 5, 46), cftime.DatetimeNoLeap(2004, 3, 16, 12, 0, 0, 0, 6, 75), cftime.DatetimeNoLeap(2004, 4, 16, 0, 0, 0, 0, 2, 106), cftime.DatetimeNoLeap(2004, 5, 16, 12, 0, 0, 0, 4, 136), cftime.DatetimeNoLeap(2004, 6, 16, 0, 0, 0, 0, 0, 167), cftime.DatetimeNoLeap(2004, 7, 16, 12, 0, 0, 0, 2, 197), cftime.DatetimeNoLeap(2004, 8, 16, 12, 0, 0, 0, 5, 228), cftime.DatetimeNoLeap(2004, 9, 16, 0, 0, 0, 0, 1, 259), cftime.DatetimeNoLeap(2004, 10, 16, 12, 0, 0, 0, 3, 289), cftime.DatetimeNoLeap(2004, 11, 16, 0, 0, 0, 0, 6, 320), cftime.DatetimeNoLeap(2004, 12, 16, 12, 0, 0, 0, 1, 350), cftime.DatetimeNoLeap(2005, 1, 16, 12, 0, 0, 0, 4, 16), cftime.DatetimeNoLeap(2005, 2, 15, 0, 0, 0, 0, 6, 46), cftime.DatetimeNoLeap(2005, 3, 16, 12, 0, 0, 0, 0, 75), cftime.DatetimeNoLeap(2005, 4, 16, 0, 0, 0, 0, 3, 106), cftime.DatetimeNoLeap(2005, 5, 16, 12, 0, 0, 0, 5, 136), cftime.DatetimeNoLeap(2005, 6, 16, 0, 0, 0, 0, 1, 167), cftime.DatetimeNoLeap(2005, 7, 16, 12, 0, 0, 0, 3, 197), cftime.DatetimeNoLeap(2005, 8, 16, 12, 0, 0, 0, 6, 228), cftime.DatetimeNoLeap(2005, 9, 16, 0, 0, 0, 0, 2, 259), cftime.DatetimeNoLeap(2005, 10, 16, 12, 0, 0, 0, 4, 289), cftime.DatetimeNoLeap(2005, 11, 16, 0, 0, 0, 0, 0, 320), cftime.DatetimeNoLeap(2005, 12, 16, 12, 0, 0, 0, 2, 350), cftime.DatetimeNoLeap(2006, 1, 16, 12, 0, 0, 0, 5, 16), cftime.DatetimeNoLeap(2006, 2, 15, 0, 0, 0, 0, 0, 46), cftime.DatetimeNoLeap(2006, 3, 16, 12, 0, 0, 0, 1, 75), cftime.DatetimeNoLeap(2006, 4, 16, 0, 0, 0, 0, 4, 106), cftime.DatetimeNoLeap(2006, 5, 16, 12, 0, 0, 0, 6, 136), cftime.DatetimeNoLeap(2006, 6, 16, 0, 0, 0, 0, 2, 167), cftime.DatetimeNoLeap(2006, 7, 16, 12, 0, 0, 0, 4, 197), cftime.DatetimeNoLeap(2006, 8, 16, 12, 0, 0, 0, 0, 228), cftime.DatetimeNoLeap(2006, 9, 16, 0, 0, 0, 0, 3, 259), cftime.DatetimeNoLeap(2006, 10, 16, 12, 0, 0, 0, 5, 289), cftime.DatetimeNoLeap(2006, 11, 16, 0, 0, 0, 0, 1, 320), cftime.DatetimeNoLeap(2006, 12, 16, 12, 0, 0, 0, 3, 350), cftime.DatetimeNoLeap(2007, 1, 16, 12, 0, 0, 0, 6, 16), cftime.DatetimeNoLeap(2007, 2, 15, 0, 0, 0, 0, 1, 46), cftime.DatetimeNoLeap(2007, 3, 16, 12, 0, 0, 0, 2, 75), cftime.DatetimeNoLeap(2007, 4, 16, 0, 0, 0, 0, 5, 106), cftime.DatetimeNoLeap(2007, 5, 16, 12, 0, 0, 0, 0, 136), cftime.DatetimeNoLeap(2007, 6, 16, 0, 0, 0, 0, 3, 167), cftime.DatetimeNoLeap(2007, 7, 16, 12, 0, 0, 0, 5, 197), cftime.DatetimeNoLeap(2007, 8, 16, 12, 0, 0, 0, 1, 228), cftime.DatetimeNoLeap(2007, 9, 16, 0, 0, 0, 0, 4, 259), cftime.DatetimeNoLeap(2007, 10, 16, 12, 0, 0, 0, 6, 289), cftime.DatetimeNoLeap(2007, 11, 16, 0, 0, 0, 0, 2, 320), cftime.DatetimeNoLeap(2007, 12, 16, 12, 0, 0, 0, 4, 350)], dtype=object)
- Coriolis(yq, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Coriolis parameter at corner (Bu) points
- units :
- s-1
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - areacello(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Ocean Grid-Cell Area
- units :
- m2
- cell_methods :
- area:sum yh:sum xh:sum time: point
- standard_name :
- cell_area
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - areacello_bu(yq, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Ocean Grid-Cell Area
- units :
- m2
- cell_methods :
- area:sum yq:sum xq:sum time: point
- standard_name :
- cell_area
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - areacello_cu(yh, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Ocean Grid-Cell Area
- units :
- m2
- cell_methods :
- area:sum yh:sum xq:sum time: point
- standard_name :
- cell_area
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - areacello_cv(yq, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Ocean Grid-Cell Area
- units :
- m2
- cell_methods :
- area:sum yq:sum xh:sum time: point
- standard_name :
- cell_area
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - deptho(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Sea Floor Depth
- units :
- m
- cell_methods :
- area:mean yh:mean xh:mean time: point
- cell_measures :
- area: areacello
- standard_name :
- sea_floor_depth_below_geoid
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - dxCu(yh, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Delta(x) at u points (meter)
- units :
- m
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - dxCv(yq, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Delta(x) at v points (meter)
- units :
- m
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - dxt(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Delta(x) at thickness/tracer points (meter)
- units :
- m
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - dyCu(yh, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Delta(y) at u points (meter)
- units :
- m
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - dyCv(yq, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Delta(y) at v points (meter)
- units :
- m
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - dyt(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Delta(y) at thickness/tracer points (meter)
- units :
- m
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolat(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Latitude of tracer (T) points
- units :
- degrees_north
- cell_methods :
- time: point
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolat_c(yq, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Latitude of corner (Bu) points
- units :
- degrees_north
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolat_u(yh, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Latitude of zonal velocity (Cu) points
- units :
- degrees_north
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolat_v(yq, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Latitude of meridional velocity (Cv) points
- units :
- degrees_north
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolon(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Longitude of tracer (T) points
- units :
- degrees_east
- cell_methods :
- time: point
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolon_c(yq, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Longitude of corner (Bu) points
- units :
- degrees_east
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolon_u(yh, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Longitude of zonal velocity (Cu) points
- units :
- degrees_east
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - geolon_v(yq, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Longitude of meridional velocity (Cv) points
- units :
- degrees_east
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - hfgeou(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Upward geothermal heat flux at sea floor
- units :
- W m-2
- cell_methods :
- area:mean yh:mean xh:mean time: point
- standard_name :
- upward_geothermal_heat_flux_at_sea_floor
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - sftof(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- Sea Area Fraction
- units :
- %
- cell_methods :
- area:mean yh:mean xh:mean time: point
- standard_name :
- SeaAreaFraction
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - thkcello(z_l, yh, xh)float32dask.array<chunksize=(1, 576, 720), meta=np.ndarray>
- id :
- thkcello
- name :
- thkcello
- units :
- m
- long_name :
- ocean model cell thickness
- original_name :
- cell_thickness
- standard_name :
- cell_thickness
- comment :
- thkcello is the nominal cell thickness in z* coordinates. The model actual thkcello is time-dependent and can be calculated as thkcello * ( deptho + zos ) / deptho
Array Chunk Bytes 58.06 MB 1.66 MB Shape (35, 576, 720) (1, 576, 720) Count 36 Tasks 35 Chunks Type float32 numpy.ndarray - wet(yh, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- 0 if land, 1 if ocean at tracer points
- units :
- none
- cell_methods :
- time: point
- cell_measures :
- area: areacello
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - wet_c(yq, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- 0 if land, 1 if ocean at corner (Bu) points
- units :
- none
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - wet_u(yh, xq)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- 0 if land, 1 if ocean at zonal velocity (Cu) points
- units :
- none
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - wet_v(yq, xh)float32dask.array<chunksize=(576, 720), meta=np.ndarray>
- long_name :
- 0 if land, 1 if ocean at meridional velocity (Cv) points
- units :
- none
- cell_methods :
- time: point
- interp_method :
- none
Array Chunk Bytes 1.66 MB 1.66 MB Shape (576, 720) (576, 720) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - average_DT(time)timedelta64[ns]dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- Length of average period
Array Chunk Bytes 480 B 8 B Shape (60,) (1,) Count 61 Tasks 60 Chunks Type timedelta64[ns] numpy.ndarray - average_T1(time)datetime64[ns]dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- Start time for average period
Array Chunk Bytes 480 B 8 B Shape (60,) (1,) Count 61 Tasks 60 Chunks Type datetime64[ns] numpy.ndarray - average_T2(time)datetime64[ns]dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- End time for average period
Array Chunk Bytes 480 B 8 B Shape (60,) (1,) Count 61 Tasks 60 Chunks Type datetime64[ns] numpy.ndarray - so(time, z_l, yh, xh)float32dask.array<chunksize=(1, 1, 576, 720), meta=np.ndarray>
- long_name :
- Sea Water Salinity
- units :
- psu
- cell_methods :
- area:mean z_l:mean yh:mean xh:mean time: mean
- cell_measures :
- volume: volcello area: areacello
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- sea_water_salinity
Array Chunk Bytes 3.48 GB 1.66 MB Shape (60, 35, 576, 720) (1, 1, 576, 720) Count 2101 Tasks 2100 Chunks Type float32 numpy.ndarray - time_bnds(time, nv)timedelta64[ns]dask.array<chunksize=(1, 2), meta=np.ndarray>
- long_name :
- time axis boundaries
- calendar :
- NOLEAP
Array Chunk Bytes 960 B 16 B Shape (60, 2) (1, 2) Count 61 Tasks 60 Chunks Type timedelta64[ns] numpy.ndarray - thetao(time, z_l, yh, xh)float32dask.array<chunksize=(1, 1, 576, 720), meta=np.ndarray>
- long_name :
- Sea Water Potential Temperature
- units :
- degC
- cell_methods :
- area:mean z_l:mean yh:mean xh:mean time: mean
- cell_measures :
- volume: volcello area: areacello
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- sea_water_potential_temperature
Array Chunk Bytes 3.48 GB 1.66 MB Shape (60, 35, 576, 720) (1, 1, 576, 720) Count 2101 Tasks 2100 Chunks Type float32 numpy.ndarray - umo(time, z_l, yh, xq)float32dask.array<chunksize=(1, 1, 576, 720), meta=np.ndarray>
- long_name :
- Ocean Mass X Transport
- units :
- kg s-1
- cell_methods :
- z_l:sum yh:sum xq:point time: mean
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- ocean_mass_x_transport
- interp_method :
- none
Array Chunk Bytes 3.48 GB 1.66 MB Shape (60, 35, 576, 720) (1, 1, 576, 720) Count 2101 Tasks 2100 Chunks Type float32 numpy.ndarray - uo(time, z_l, yh, xq)float32dask.array<chunksize=(1, 1, 576, 720), meta=np.ndarray>
- long_name :
- Sea Water X Velocity
- units :
- m s-1
- cell_methods :
- z_l:mean yh:mean xq:point time: mean
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- sea_water_x_velocity
- interp_method :
- none
Array Chunk Bytes 3.48 GB 1.66 MB Shape (60, 35, 576, 720) (1, 1, 576, 720) Count 2101 Tasks 2100 Chunks Type float32 numpy.ndarray - vmo(time, z_l, yq, xh)float32dask.array<chunksize=(1, 1, 576, 720), meta=np.ndarray>
- long_name :
- Ocean Mass Y Transport
- units :
- kg s-1
- cell_methods :
- z_l:sum yq:point xh:sum time: mean
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- ocean_mass_y_transport
- interp_method :
- none
Array Chunk Bytes 3.48 GB 1.66 MB Shape (60, 35, 576, 720) (1, 1, 576, 720) Count 2101 Tasks 2100 Chunks Type float32 numpy.ndarray - vo(time, z_l, yq, xh)float32dask.array<chunksize=(1, 1, 576, 720), meta=np.ndarray>
- long_name :
- Sea Water Y Velocity
- units :
- m s-1
- cell_methods :
- z_l:mean yq:point xh:mean time: mean
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- sea_water_y_velocity
- interp_method :
- none
Array Chunk Bytes 3.48 GB 1.66 MB Shape (60, 35, 576, 720) (1, 1, 576, 720) Count 2101 Tasks 2100 Chunks Type float32 numpy.ndarray - volcello(time, z_l, yh, xh)float32dask.array<chunksize=(1, 1, 576, 720), meta=np.ndarray>
- long_name :
- Ocean grid-cell volume
- units :
- m3
- cell_methods :
- area:sum z_l:sum yh:sum xh:sum time: mean
- cell_measures :
- area: areacello
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- ocean_volume
Array Chunk Bytes 3.48 GB 1.66 MB Shape (60, 35, 576, 720) (1, 1, 576, 720) Count 2101 Tasks 2100 Chunks Type float32 numpy.ndarray - zos(time, yh, xh)float32dask.array<chunksize=(1, 576, 720), meta=np.ndarray>
- long_name :
- Sea surface height above geoid
- units :
- m
- cell_methods :
- area:mean yh:mean xh:mean time: mean
- cell_measures :
- area: areacello
- time_avg_info :
- average_T1,average_T2,average_DT
- standard_name :
- sea_surface_height_above_geoid
Array Chunk Bytes 99.53 MB 1.66 MB Shape (60, 576, 720) (1, 576, 720) Count 61 Tasks 60 Chunks Type float32 numpy.ndarray
- filename :
- ocean_monthly.200301-200712.zos.nc
- title :
- OM4p5_IAF_BLING_CFC_abio_csf_mle200
- associated_files :
- areacello: 20030101.ocean_static.nc
- grid_type :
- regular
- grid_tile :
- N/A
- external_variables :
- areacello
2D horizontal averaging
Probably one of the most used data reduction. Since the ocean grid is not uniform, we need to weight by the cell area. Special care should also be taken with land values as we’re gonna see next. The horizontal averaging can then be computed :
\(\large \overline{tracer} = \Sigma_{i,j} (tracer ~\times~ dx ~\times~ dy ~\times~ mask) ~ / ~ \Sigma_{i,j} (dx ~\times~ dy ~\times~ mask)\)
where mask is a binary mask (0: land, 1:ocean). In MOM6 terminology, this would be written (for a tracer) as:
\(\large \overline{tracer} = \Sigma_{i,j} (tracer ~\times~ areacello ~\times~ wet) ~ / ~ \Sigma_{i,j} (areacello ~\times~ wet)\)
NB: \(\large areacello \neq dxt * dyt\), with error of a couple percent, as we can see here:
[6]:
area_diff = (ds['dxt'].fillna(0.) * ds['dyt'].fillna(0.) * ds['wet'].fillna(0.)) - \
(ds['areacello'].fillna(0.) * ds['wet'].fillna(0.))
area_error = 100 * area_diff / ds['areacello']
area_error.plot()
[6]:
<matplotlib.collections.QuadMesh at 0x2b1d8606f630>

[7]:
def horizontal_mean(da, metrics):
num = (da * metrics['areacello'] * metrics['wet']).sum(dim=['xh', 'yh'])
denom = (metrics['areacello'] * metrics['wet']).sum(dim=['xh', 'yh'])
return num / denom
[8]:
sst = ds['thetao'].isel(z_l=0)
mean_sst = horizontal_mean(sst, ds)
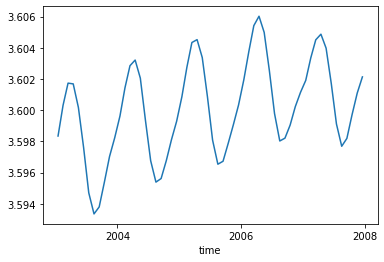
For reference, we print the first value of the obtained time-serie:
[9]:
mean_sst.isel(time=0).values
[9]:
array(18.483639, dtype=float32)
If you don’t have the “wet” array, you can infer the land/sea mask using:
[10]:
lsm = ~np.isnan(sst.isel(time=0))
and define a new function:
[11]:
def horizontal_mean_no_wet(da, metrics, lsm):
num = (da * metrics['areacello']).sum(dim=['xh', 'yh'])
denom = (metrics['areacello'].where(lsm)).sum(dim=['xh', 'yh'])
return num / denom
We verify that we get the same answer:
[12]:
mean_sst = horizontal_mean_no_wet(sst, ds, lsm)
mean_sst.isel(time=0).values
[12]:
array(18.483639, dtype=float32)
[13]:
mean_sst.plot()
[13]:
[<matplotlib.lines.Line2D at 0x2b1d87abd7b8>]
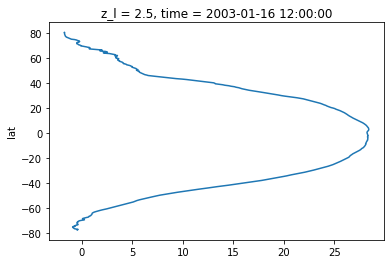
Zonal average
Zonal average is just a simpler case, where averaging is only done over the longitude and can be written as:
\(\large \overline{tracer} = \Sigma_{i} (tracer ~\times~ dxt ~\times~ wet) ~ / ~ \Sigma_{x} (dxt ~\times~ wet)\)
and we can define the function as:
[14]:
def zonal_mean(da, metrics):
num = (da * metrics['dxt'] * metrics['wet']).sum(dim=['xh'])
denom = (metrics['dxt'] * metrics['wet']).sum(dim=['xh'])
return num/denom
[15]:
zonalmean_sst = zonal_mean(sst, ds)
If we want to plot the zonal mean, we also need to have the correct latitude array and not the so-called nominal coordinate yh. We can calculate it with the same formula:
[16]:
correct_lat = zonal_mean(ds['geolat'], ds)
Then replace the bad latitudes with the correct ones in the data array:
[17]:
zonalmean_sst = zonalmean_sst.rename({'yh': 'lat'})
zonalmean_sst['lat'] = correct_lat.values
[18]:
zonalmean_sst.isel(time=0).plot(y='lat')
[18]:
[<matplotlib.lines.Line2D at 0x2b1d87b71518>]
3D average
Same method than for the 2D horizontal average, except we use volume instead of area and sum over 3 dimensions:
\(\large \overline{tracer} = \Sigma_{i,j,k} (tracer ~\times~ dx ~\times~ dy ~\times~ dz ~\times~ mask) ~ / ~ \Sigma_{i,j,k} (dx ~\times~ dy ~\times~ dz ~\times~ mask)\)
or in MOM6 terminology, this would be written (for a tracer) as:
\(\large \overline{tracer} = \Sigma_{i,j,k} (tracer ~\times~ volcello ~\times~ wet) ~ / ~ \Sigma_{i,j,k} (volcello ~\times~ wet)\)
the function is:
[19]:
def global_mean(da, metrics):
num = (da.fillna(0.) * metrics['volcello'].fillna(0.)).sum(dim=['xh', 'yh', 'z_l'])
denom = (metrics['volcello'].fillna(0.)).sum(dim=['xh', 'yh', 'z_l'])
return num / denom
[20]:
temp_global = global_mean(ds['thetao'], ds)
[21]:
temp_global.isel(time=10).values
[21]:
array(3.5970294, dtype=float32)
Let’s make the compute go a little faster with dask.
[22]:
do_full_computation = True
if do_full_computation:
from dask.distributed import Client
from dask.distributed import LocalCluster
cluster = LocalCluster()
client = Client(cluster)
client
[23]:
if do_full_computation:
temp_global.plot()
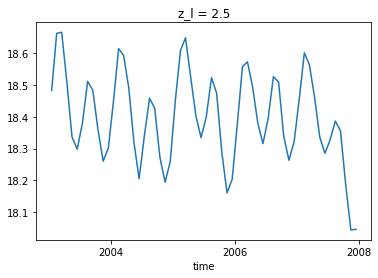
Using xgcm
xgcm provides some functions that perform the same computations in a elegant way. In order to get the right answers with MOM6, we need to pay particular attention to which metrics we’re using. Let’s show an example:
[24]:
from xgcm import Grid
We are pretty confident the function we wrote gives the right answer so we take it as our truth:
[25]:
sst = ds['thetao'].isel(z_l=0)
mean_sst = horizontal_mean(sst, ds)
mean_sst.isel(time=0).values
[25]:
array(18.483639, dtype=float32)
Now let’s define the xgcm grid with only \(dxt\) and \(dyt\):
[26]:
# make sure to fill in nans with zeros
ds['dxt'] = ds['dxt'].fillna(0.)
ds['dyt'] = ds['dyt'].fillna(0.)
metrics = {
('X',): ['dxt'], # X distances
('Y',): ['dyt'] # Y distances
}
coords={'X': {'center': 'xh', 'right': 'xq'},
'Y': {'center': 'yh', 'right': 'yq'} }
grid = Grid(ds, coords=coords, metrics=metrics, periodic=['X'])
[27]:
mean_sst_xgcm = grid.average(sst, ['X', 'Y'])
mean_sst_xgcm.isel(time=0).values
[27]:
array(18.482277, dtype=float32)
Ugh not good… Third decimal is off, but remember we need to use areacello, not dxt * dyt:
[28]:
ds['areacello'] = ds['areacello'].fillna(0.)
metrics = {
('X',): ['dxt'], # X distances
('Y',): ['dyt'], # Y distances
('X', 'Y'): ['areacello'] # Areas
}
coords={'X': {'center': 'xh', 'right': 'xq'},
'Y': {'center': 'yh', 'right': 'yq'} }
grid = Grid(ds, coords=coords, metrics=metrics, periodic=['X'])
[29]:
mean_sst_xgcm = grid.average(sst, ['X', 'Y'])
mean_sst_xgcm.isel(time=0).values
[29]:
array(18.483637, dtype=float32)
Now we’re consistent to the 6th decimal point.
Now with the 3d averaging, we can define a nominal layer thickness as:
[30]:
ds['dzt'] = xr.DataArray(data=ds['z_i'].diff('z_i').values,
coords={'z_l': ds['z_l']},
dims=('z_l'))
However, because of the partial steps the volume of the cell can be different from areacello * dzt so we need to add volcello to the metrics:
[31]:
ds['volcello'] = ds['volcello'].fillna(0.)
metrics = {
('X',): ['dxt'], # X distances
('Y',): ['dyt'], # Y distances
('Z',): ['dzt'],
('X', 'Y'): ['areacello'], # Areas
('X', 'Y', 'Z'): ['volcello'], # Volumes
}
coords={'X': {'center': 'xh', 'right': 'xq'},
'Y': {'center': 'yh', 'right': 'yq'},
'Z': {'center': 'z_l', 'outer': 'z_i'}}
grid = Grid(ds, coords=coords, metrics=metrics, periodic=['X'])
[32]:
%%time
mean_temp_xgcm = grid.average(ds['thetao'], ['X', 'Y', 'Z'])
# this is a bit long...
CPU times: user 18.1 s, sys: 413 ms, total: 18.5 s
Wall time: 51.6 s
[33]:
mean_temp_xgcm.isel(time=10).values
[33]:
array(3.597029, dtype=float32)
Which is in good agreement with our global_mean function!
[34]:
if do_full_computation:
cluster.close()
client.close()
[ ]: